Data Download - Obtain & Preprocess the Links
Introduction
Before files can be downloaded, we need to extract the links we need. For this section, we have three components: obtaining download links, removing unwanted Entries, and extracting to file list.
Obtaining Download Links
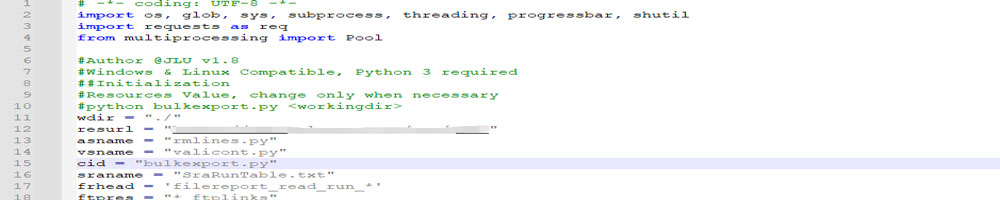
For each run accession number(SRRXXXXXXX), two files were obtained: one SraRunTable.txt and filereport_read_run_PRJNAXXXXX_tsv.txt. The accession number was obtained via SRA's online searches with specific keywords. SraRunTable.txt was also downloaded via SRA web. While downloading, only column "Run" and "Assay Type" was selected.
The other file was obtained using the accession number via ENA EBI's website. Unlike SRA, ENA provides direct FTP access, which will make the downloading process easier.
We manually set the parameters while searching. Once the URL was generated, a Python script was used to automatically download all the metadata. The result is 277,592 accessions waiting to be downloaded.
Remove Unwanted Entries
Based on the researchers' requirement, we only want the entries where the "Assay Type" is "RNA-Seq". In addition, we only want the result of three species: "Homo sapiens", "Mus Musculus", and "Rattus norvegicus".
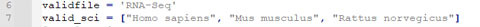
Extracting to File List
Assigning Download Batches
Each accession ranges from 2 to 20,000 download links. For ease of management, we have to download multiple accessions together. Therefore I choose to assign every 50 accessions to one batch.
Obtaining Downloading List
Once each file was processed and only the entries we wanted were left, another Python script called "bulkexport.py" was applied to each batch to extract all download links from its content to one file called "_url".
Once we have this list of URLs, it can be passed to downloading servers.
Navigate through the Data Download Section
- Prev
- Overview
- Obtain & Preprocess the Links
- Software & Structure
- Testing & Firewall Circumvention
- Error Processing
- Next