Data Process - Metadata Storage & Database

Introduction
Managing thousands of files each day can be complex, and I plan to create a database to store the metadata for all files/accessions/studies.
File Structure
Each accession ("SRXXXXX") belongs to a study ("PRJNAXXXX") and each accession has a list of gene sequences ("SRRXXXXX.fastq.gz"). The metadata was stored with each accession.
List of Metadata:
| File Name | Content / Purpose | Source |
|---|---|---|
| filereport_read_run_PRJNAXXXXX_tsv.txt | Metadata contains downloading links. | ENA Website |
| SraRunTable.txt | Contains Assay Type of accession | SRA Website |
| SRPXXXXX_content_XX | File contains all files with in each accession | Script: bulkexport.py |
| SRPXXXXX_ftplinks | Contains all ftp links. | Script: bulkexport.py |
| .ContentValidated | A mark of a downloaded accession. | Script: movef.py |
Database
Content
The database should be separated into two relations. One for each file, which contains its size, name, and accession belongs to. One for each accession, which contains the number and name of files contained, the study name, and physical/volume identifiers for storage. In addition, each section should contain the creation and last update time and user, since multiple users are using this system.
Table
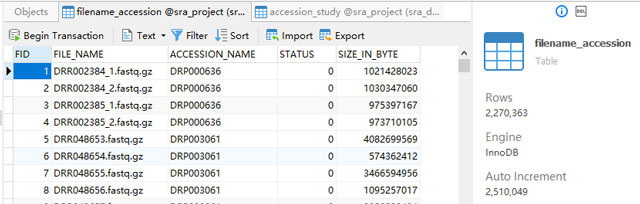
Based on the requirement, two tables were created. One called "filename_accession" which stores information about each file and maps its relation to the accession. The other one is "accession_study", which stores information about each study and maps its relation to the study name. We don't have much information for each study other than its name, so I didn't create a table for the study.
Update
I have created a script "update_information.py" that updates information for contents in each accession directory. During each step of our operation, this script was first invoked to have the status reflected in the database.
Columns
Due to security reasons, the complete columns or table creation SQL was not shown.
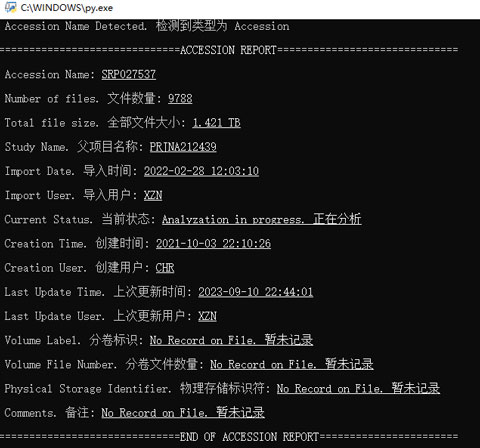
Query
The database can also be used to inquire data when needed. I have developed a query script as an interface for users to inquire about the database, which can automatically recognize the input string and query with appropriate SQL. Also, the script will show additional information like the total file size calculated based on the raw data retrieved from the database.