Data Analysis - Software & Structure
Without using platforms like Galaxy, I have to build my own analysis pipeline using Python. The main idea is to use Python to manage files and invoke the software used by each step. It serves as a framework where each component (software invoked) can be replaced or updated at any time.
This section has two components: software & dependencies and structure.

Software & Dependencies
Based on the pipeline created by the researchers, there are three software used: fastqc, trim_galore, and salmon. I don't really understand the purpose of this software, as they are used to process and analyze gene sequence. However, I can still create the pipeline using the predefined parameters.
The software requires reference genes to run, so the reference genes of the three species were downloaded. Additionally, bbmap was used to test and convert the file format(from phred64 to phred33).
I have put the software and dependencies in the exact same location for all three analysis servers. This will ease our maintenance in the future.

Structure
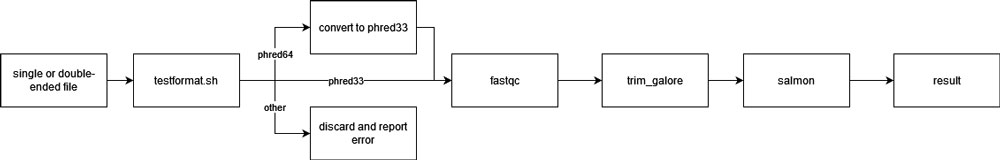
The gene sequence was processed in the unit of single files. That is, each file (or pair of interleaved-ended files) will be processed individually and independently. For each file, it will first be tested to see if it is phred33 or phred64 format. If the file is in phred64 format, it will be converted into phred33. If an unsupported format or unpaired interleaved-ended file is detected, the file will be discarded. Once the file is OK, the scientific name will be acquired from the metadata and the corresponding reference will be passed to fastqc, trim_galore, and finally salmon. Once the salmon is finished running, the result is generated. The result for each file is then collected and combined once the whole accession is finished running.
Since the file processed might be converted to phred33 format, we want to store the processed file instead of the original. In that case, we won't need to process the file again if it is needed in the future. It is possible that in the future the original format will be preferred, but for now, we have decided to store in phred33 format after discussion.
Navigate through the Data Analysis Section
- Prev
- Overview
- Software & Structure
- Optimization & Performance Improvement
- Dealing with Errors
- Result Collection
- Automation
- Next