Preliminary - Performance Estimation
Introduction
Once we have settled the initial software, it is time to estimate the amount of performance needed. In this section, only performance-related issues like CPU, network, and bandwidth will be discussed. For things like the number of hard drives, please refer to the next section - resource estimation.
This section contains five parts: Total Amount of Data, Downloading Bandwidth, Internal Network(LAN), CPU & Memory Cost, and Storage Plan.
Total Amount of Data
To estimate the amount of data, we need to know which kind of data we need. Based on the filter that researchers provided, I will be able to obtain the list of files we need from the SRA website. The next step is to obtain the metadata of each file and calculate the total amount of data. To do this, I write a Python crawler to iteratively pull information from the SRA website and then calculate the sum of all sizes. To make things easier, I provided one of the researchers with this script and asked him to apply his filter and then collect the result. (This has been proved to be a mistake later.)
To avoid being banned by the firewall, it took 4 days to have all the metadata downloaded. There is a total of ~430 TB in the end.
In a later stage when I developed the analysis script, I re-ran everything and found the data should be 277,592 accessions for a total of ~2000 TB. However, for now, we will stick with 430TB for the estimations.
Downloading bandwidth
There are a total of 430TB of data, and it should be good to have the project finished in 2 - 3 years. Based on these statistics, we should have 150 TB downloaded each year, about 500 - 700 GB per day.
700 GB / day = ~29 GB / hr = ~495 MB / min = 8.24 MB / s = ~66 Mbps/
Considering the complex network conditions in China, a 500 Mbps line should be good for now. This later expands to two 1 Gbps lines with the establishment of my mini data center. For more information on it, please refers to the datacenter - network project.

Internal Network(LAN)
Unlike the external bandwidth that downloads 24/7, LAN needs to be robust and fast. For a file that is 8 GB, it will take the conventional 1000M link about 1 minute to transfer at the maximum theoretical speed (125MB/s). Imagine the server needs to wait for 1 minute just to fetch one of thousands of files from the storage nodes. In addition, it is very common for gene sequences to be over 10 GB.
We need something faster than 1G. For enterprise-grade equipment, the next level of speed is usually 10 GB on an SFP+ fiber link or CAT 8 cable. 10GB/s should be fast enough in most cases. It takes about 6 seconds to transfer a file of 8 GB. With a 10 GB link, the performance is more likely to be bottlenecked by the disk performance than the network.
I eventually go for a 10G/40G hybrid network, which will be discussed later.
CPU & Memory Cost
The Galaxy Platform
I start by trying Galaxy. First, a virtual machine was created. This VM has 16 dedicated cores (From Intel Xeoon E7-8895 v2) and 128 GB DDR3 Memory. I estimate this should be enough for a performance test. It took me some time to have the Galaxy platform set up on our own VM. It took another week for researchers to create a workable pipeline on the platform.

After testing, It took 12 hours for Galaxy to process a 1.3G file with 1 core CPU and 3 GB memory. This is really slow. What makes things worse, Galaxy requires full GUI operation. which means human interaction is needed to process each file. Given the processing time for each file varies a lot and we need the server to run 24/7, using Galaxy is not practical!
However, back in time, the researchers told me that they could handle the manual part and that I didn't need to worry about this. For simplicity, I include the final plan that I actually used below. But for now, we will assume using Galaxy for the rest parts of the estimation section.
The Final Plan - CPU
Get rid of the Galaxy, I'm now on my own.
First, I collected the software and parameters used in the pipeline that researchers created. Second, I manually downloaded the software and tried to process a file manually. There are a total of 4 components needed: fastqc, trim_galore, salmon, and reference genes for 3 species.
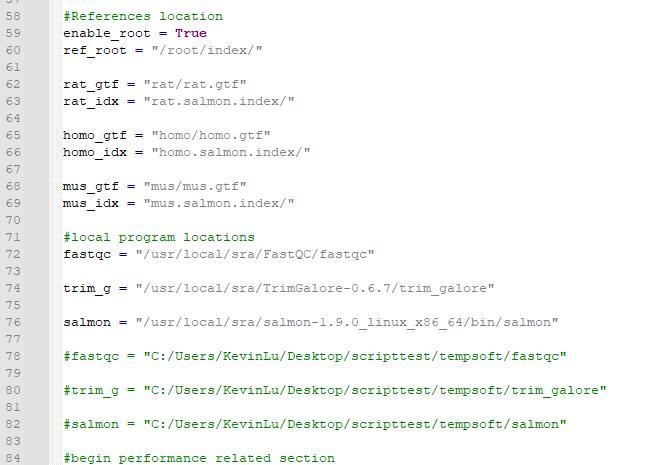
Luckily, I was able to manually create a pipeline using the above software/files. During the testing, I found a maximum of 7 threads should be used for a single file (or file pair for interleaved-ended sequences). In addition, pigz should be installed so that it will take Salmon less time on file compression.
Although I couldn't remember the exact technical details by the time I wrote this post, the new pipeline is over 5 times faster than the Galaxy options. In addition, all the components are command-line utilities and thus could be fully automated using Python script.
The Final Plan - Memory
The memory requirement is also much less than the Galaxy options. I couldn't notice significant memory usage during the test, and at least 32 GB for the analysis server each with ~104 logical cores should be more than enough.
Storage Plan
Before choosing storage backends, I need to figure out which data needs to be stored. After a discussion with the researchers, we decided to store the analysis result (of course), and as much source data as we can.
Usually, for data storage, we have 4 options: HDD, SSD, CD/DVD, and Tape.
| Meterial | Advantage | Disadvantage |
|---|---|---|
| HDD | Quick access, can be read by any device. | Expensive for large amount of data. |
| SSD | Extremely quick access, small in size. | Extremely expensive, potential data lost if not powered on for a long time. |
| CD/DVD | Cheap, and can be read by common devices. | Capacity is very limited for each CD |
| Tape | Relatively cheap for large-scale data storage. Fast R/W (sequential) | Tape drives can be very expensive, no random file access. |
For our purposes, using SSD is apparently not appropriate due to its cost and problem with long-term storage. Using CD/DVD is also not practical due to its capacity. I have to decide between HDD and Tape.
After thorough research and comparing the price difference, I found the tape library (LTO6) will supersede the HDD options once the total amount of data exceeds ~330 TB. In our situation, using tape is a good choice.

However, given the poor performance of tape, it should only be used for cold data storage. For those data that are still downloading or in the process of analysis, HDDs are still needed. That is, I need a hybrid storage backend that contains both HDD and Tape.